 We all share a concern for Antimicrobial Resistance (AMR) and its impact on human medicine, veterinary medicine, and global society. As veterinarians, our first instinct is to correct the problem! As scientists, we want data to help craft the plan. As a mother (and as a human being in general), I (and others) want to do what is right. The best approach is often a combination of all three—quick action to do what is right-driven by data to support. The concern with AMR is no different.
We all share a concern for Antimicrobial Resistance (AMR) and its impact on human medicine, veterinary medicine, and global society. As veterinarians, our first instinct is to correct the problem! As scientists, we want data to help craft the plan. As a mother (and as a human being in general), I (and others) want to do what is right. The best approach is often a combination of all three—quick action to do what is right-driven by data to support. The concern with AMR is no different.
AMR refers to a bacteria’s ability to be killed by an antibiotic. The bacteria that survives an antibiotic regimen are considered resistant and can cause illnesses that can be difficult to treat. It seems so simple, but can be so complex. There still so much to be learned.
What we DO Know:
AMR has been around for thousands of years, even before antibiotics were commercially available. Some antibiotics were developed from naturally occurring compounds in the environment (think Penicillin developed from mold). AMR developed initially as a defense mechanism to these naturally occurring compounds.
Once antibiotics became commercially available in the 1950’s, the pressure on bacterial populations changed. This inadvertently creates AMR, even when used responsibly.
We can detect antibiotic resistance in bacteria from humans, animals, food and the environment (water, air, soil).
AMR within bacteria can be passed on to the next generation of bacteria. Additionally, AMR and genetic material can be shared with other bacteria. So generous, isn’t it?
To better understand AMR and how it is changing in humans, animals, and the environment, most countries have started monitoring bacteria. In the United States, the National Antimicrobial Resistance Monitoring System (NARMS) monitors enteric bacteria (aka from the gut) of sick people, from meat at the grocery stores, and samples at the meatpacking plants. Significant amounts of data and fun interactive tools are available on the NARMS website if you want to check it out to learn more!
What about the farm?
Even though agriculture is often considered a source of AMR in humans, we have not been able to find the data to know where or how to start impacting. Remember, NARMS focuses on grocery stores and packing plants, NOT animals on the farm. We immediately identified a gap and have developed a plan to take action to fill the missing void.
What we are doing to fill the knowledge gap:
Pipestone is working on addressing this knowledge gap through a few unique and exciting projects:
- Tracking Antibiotic Resistance Over Time from Diagnostics:
We took 17 years of diagnostic cases submitted by Pipestone veterinarians to the University of Minnesota, Iowa State University, and South Dakota State University and plotted the resistance over time. From this, we have learned that despite the use of antimicrobials over a prolonged period, the overall level of resistance has not changed. We continue to update and publish this graph monthly on www.PipestonePART.com .
The graph below depicts information regarding antibiotic resistance across 17 years. 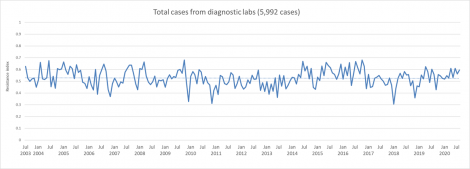
The red line represents the annual resistance index from 2003 to 2020.
The resistance index ranges from 0 to 1 with 0 (no resistance) and 1 (all cases submitted were resistant).
The blue bars represent the total number of cases submitted by the Pipestone Veterinary Services team each year.
Alternative Site Comparison Project:
We collected monthly samples across four different environments – a pig barn, a wastewater treatment plant, a playground, and a companion animal facility to see how different, or similar, the AMR findings were from a pig farm. Metrics evaluated included frequency of bacteria recovered and percentage of AMR. Spoiler alert: We found AMR in all sampled environments, suggesting it IS naturally occurring and not necessarily indicative of antibiotic use.
- IMAGINE Project:
This year, we began collecting diagnostic samples at commercial pig farms from both sick and healthy animals. Resistance testing will be conducted and correlated with each farm’s antibiotic use data. Because it’s the first of its kind and we as scientists need accurate data to support action. We will do this over an extended period and in different geographies across the U.S. NARMS and the International Consortium for Antimicrobial Stewardship in Agriculture (ICASA) funded through the President’s Advisory Council have provided match funds for this project. This project will serve as a model to advance the U.S. swine industry to the global forefront in areas of responsible antibiotic use and AMR surveillance.
What will we do with the data?
- Track and compare AMR for pathogens that are important to human and pig health.
- Determine AMR patterns to actual antibiotic use.
- Learn how we can impact and reduce AMR threat to our families and the pigs we care for.
IMAGINE living in a world where we know more and can use that knowledge to make a difference. Follow along on our journey to protect animal & human health and promote responsible antibiotic use on PipestonePART.com or our YouTube channel – Doing our PART.
By Dr. Carissa Odland Pipestone Veterinary Services
Sources:
Bhullar K, Waglechner N, Pawlowski A, Koteva K, Banks ED, et al. (2012) Antibiotic Resistance Is Prevalent in an Isolated Cave Microbiome. PLoS ONE 7(4): e34953. doi:10.1371/journal.pone.0034953
Van Goethem et al. “A reservoir of ‘historical’ antibiotic resistance genes in remote pristine Antarctic soils” Microbiome (2018) 6:40 https://doi.org/10.1186/s40168-018-0424-5
Aminov et al. “A brief history of the antibiotic era: lessons learned and challenges for the future” (2010) Frontiers in Microbiology 1:134 doi:10.3389/fmicb.2010.00134
Zhou and Wang. “Characteristics of antibiotic resistance of airborne Staphylococcus isolated from metro stations” Int. J. Environ. Res. Public Health 2013, 10, 2412-2426; doi:10.3390/ijerph10062412
Barrios et al. “Fate and transport of antibiotics and antibiotic resistance genes in runoff and soil as affected by the timing of swine manure slurry application” (2020). Civil and Environmental Engineering Faculty Publications. 198. https://digitalcommons.unl. edu/civilengfacpub/198
Source: CDC. National Antimicrobial Resistance Monitoring System (NARMS) Now: Human Data. Atlanta, Georgia: U.S. Department of Health and Human Services, CDC. 08/12/2020. https://www.cdc.gov/narmsnow. Accessed 8/13/2020.
